![[Course icon goes here]](icon-course-1.jpg)
CBE 255. Introduction to Chemical Process Modeling
| CBE Faculty |
| Department of Chemical and Biological Engineering |
| University of Wisconsin |
| Madison, Wisconsin |
Copyright (C) 2014 Department of Chemical and Biological Engineering
Course Overview
This course provides an overview of the chemical engineering
curriculum and develops facility with using modern computational
software for numerical problem solving.
The course provides an integrative overview of the entire
chemical engineering curriculum. The faculty instructors for the
latter courses in the curriculum have identified the key modeling
concepts that they would like students entering their courses to be
exposed to early in their education in our department. We will select
10-12 key topics from this list and structure the course around one
key concept per week. The lectures will introduce these concepts and
the students will immediately begin to apply them in smaller
discussion sections. We will introduce computational tools motivated
by these key concepts. By the end of this course, the students will
have a set of tools that have been selected by the faculty in the
latter courses as most useful to them as instructors.
Technology is an essential component of this course. The college has
invested in classrooms with laptop projection capability and wireless
internet connections. Many engineering undergraduates bring their own
laptops to campus. The department will loan the additional laptops so
every student can bring their laptop to class. The course requires
intensive problem solving in small groups to educate the students in
how to use advanced computational tools for engineering decision
making in complex situations. We envision using undergraduates who
have mastered the course in previous semesters as the leaders for the
smaller discussion sections. The professor in charge of the course
will also cover one discussion section on a rotating basis.
The course may be broken into two parts, with the first part
taught to sophomores and the second part to juniors. In this way, the
students will find almost immediate use of all the tools they have
learned in their next semester courses.
Given the following features: (i) the course provides an integrated
view of the entire curriculum, (ii) the course instructors select the
concepts and computational tools presented, and (iii) the senior
students lead discussion sections with the junior students, we hope to
foster a learning community in the department in which using
technology to solve complex engineering problems becomes an integral
part of our students' educational experience.
Complete collections of the M-files for both Matlab and Octave
in zip or tar.gz file formats are available for download from the
following links:
- Matlab:
-
- Octave:
-
Module 1: Programming and programming languages
Programming concepts: introduction to Matlab, purpose of Matlab windows, accessing the online help system.
Video Lectures
Module 2: Stoichiometry of chemical reactions
Chemical and biological engineering concepts: chemical reactions, linear independence of reactions, reaction rates, production rates
Computational concepts: matrices, rank of a matrix, submatrices, reshaping matrices, solving least squares problems
Programming concepts: looping, conditionals, plotting, loading data from files, writing to the screen
Broader applications of these concepts: electrical circuit theory, network models, linear programming and game theory, financial models and optimization
Video Lectures
Module 3: Diffusion and heat transfer
Chemical and biological engineering concepts: heat transfer and mass diffusion, gradient and flux, thermal conductivity, heat capacity, diffusivity, dimensionless variables
Computational concepts: partial differential equations, implicit differential equations, differential-algebraic equations, orthogonal collocation, semi-infinite domains
Programming concepts: colloc program, functions, scripts, global statement, ODE solvers in Matlab, ode15i
Broader applications of these concepts: transport phenomena, rate processes, dimensional analysis

| Figure 2 (page 6):
Temperature profile at several times during heating of a slab; various kt/(\rho C_Pb^2).
|
|

| Figure 3 (page 7):
Temperature profile of the semi-infinite slab at different \tau = kt/(\rho C_P).
|
|

| Figure 5 (page 12):
Dimensionless concentration versus dimensionless radial position for different numbers of collocation points.
|
|
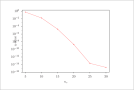
| Figure 6 (page 13):
Relative error in the effectiveness factor versus number of collocation points.
|
|
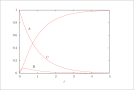
| Figure 7 (page 15):
The solution to the full model for the series reaction A -> B -> C; ODE model.
|
|
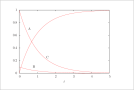
| Figure 8 (page 15):
The solution to the reduced QSSA model; DAE model.
|
|
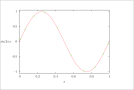
| Figure 9 (page 18):
The function sin 2 \pi x and 10 collocation points.
|
|
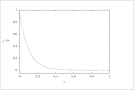
| Figure 10 (page 19):
The function e^{-10x} and 10 collocation points.
|
|
Module 4: Process systems steady-state modeling and design
Chemical and biological engineering concepts: process systems, reaction, separation, flash tank, recycle, material balances, degrees of freedom
Computational concepts: solving sets of nonlinear algebraic equations, Newton's method, Jacobian matrix
Programming concepts: looping, iteration, formatted output to the screen, user input from the keyboard, nonlinear algebraic equation solvers in Matlab

| Figure 2 (page 5):
Two iterations of Newton's method for f(x) = x^3 - 2x^2 + 3x - 6 = 0.
|
|
|
|
Video Lectures
Module 5: Chemical kinetics in well-mixed reactors
Chemical and biological engineering concepts: chemical kinetics, law of mass action, material balance, well-mixed reactor, reaction rates, production rates, complex dynamics, oscillations, Zhabotinsky reaction, coupled mass and energy balance for the CSTR
Computational concepts: ordinary differential equations, Euler method, stepsize, relative and absolute errors
Programming concepts: functions, scripts, global statement, ODE solvers in Matlab, ode15s
Broader applications of these concepts: dynamical systems, complex dynamics, oscillations, multiple steady states
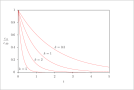
| Figure 2 (page 3):
First-order, irreversible kinetics in a batch reactor.
|
|
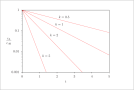
| Figure 3 (page 3):
First-order, irreversible kinetics in a batch reactor, log scale.
|
|
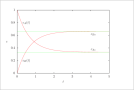
| Figure 4 (page 6):
First-order, reversible kinetics in a batch reactor, k_1=1, k_{-1}=0.5, c_{A0}=1, c_{B0}=0.
|
|

| Figure 5 (page 7):
Concentrations of species in Reactions \ref {rxn:radio1}--\ref {rxn:radio2} versus time.
|
|

| Figure 6 (page 8):
Concentrations (log scale) versus time
|
|

| Figure 8 (page 16):
Complex dynamic behavior of the Oregonator model of the Belousov-Zhabotinsky reaction.
|
|
Video Lectures
Module 6: Staged separations
Module 7: Estimating parameters from data
Chemical and biological engineering concepts: fitting chemical and biological engineering models to data
Computational concepts: optimization, least squares, statistical confidence intervals, normal and uniform distributions, sampling, mean, variance
Programming concepts: parest.m, ellipse.m, Sundials package, hist, chi2inv, sqrtm, rand, randn
Broader applications of these concepts: Decision making under uncertainty, model discrimination, statistical methods, random variables, sampling

| Figure 1 (page 3):
Normal distribution, with probability density p_\xi (x) = (1/\sqrt {2\pi \sigma ^2}) o{exp}(-(1/2) (x-m)^2/\sigma ^2).
|
|

| Figure 2 (page 4):
Multivariate normal for n=2.
|
|
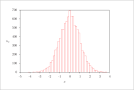
| Figure 4 (page 7):
Histogram of 10,000 samples of {\sc Matlab}'s randn function.
|
|

| Figure 5 (page 10):
The 15 samples of measurement, pressure, and concentration.
|
|
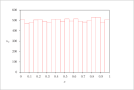
| Figure 6 (page 12):
Histogram of 10,000 samples of uniformly distributed x.
|
|
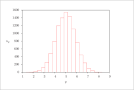
| Figure 7 (page 12):
Histogram of 10,000 samples of y= sum_{i=1}^{10} x_i.
|
|

| Figure 8 (page 14):
One thousand samples of the random variable x\sim N(m, P) and 95% probability contour.
|
|
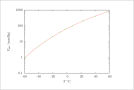
| Figure 10 (page 20):
Antoine model fitted to the acetone vapor pressure data of Example \ref {ex:AntoineFit}.
|
|

| Figure 11 (page 22):
Measurements of species concentrations in Reactions \ref {rxn:series} versus time.
|
|

| Figure 12 (page 23):
Fit of model to measurements using estimated parameters.
|
|

| Figure 13 (page 26):
Measurements of species concentrations versus time for parallel reactions.
|
|

| Figure 14 (page 31):
Measurements of species concentrations versus time for Exercise\ref {exer:ABCD}.
|
|

| Figure 15 (page 32):
Measurements of species concentrations versus time for Exercise\ref {exer:ABCD_lowB}.
|
|
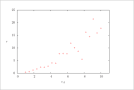
| Figure 16 (page 33):
Rate versus concentration of A.
|
|

| Figure 17 (page 35):
Concentrations of species A, C, D, and E versus time.
|
|
|
|
Video Lectures
Module 8: Course review